Summary: jq rocks for speedy JSON mangling. Use it to make powerful git clean filters, e.g. when stripping out unwanted cached-data from Jupyter notebooks. You can find the documentation of git 'clean' and 'smudge' filters buried in the page on git-attributes, or see my example setup below.
The trouble with notebooks
For a year or so now I've been using Jupyter notebooks as a means to produce tutorials and other documentation (see e.g. the voeventdb.remote tutorial). It's a powerful medium, providing a good compromise between ease-of-editing and the capability to interleave text, code, intermediate results, plots, and even nicely-typeset LaTeX-encoded equations. I've even gone to far as to urge its adoption in recent conference talks.
However, this powerful interface inherits the age-old curse of WYSIWYG editors - the document-files tend to contain more than just plain-text, and therefore are not-so-easy to handle with standard version-control tools. In the case of Jupyter, the format doesn't stray too far from comfortable plain-text territory - the ipynb format is just a custom JSON data-structure, with the occasional base-64 encoded blob for images and other binary data. Which means version-control systems such as Git can handle it quite well, but diff-comparing different versions of a complex notebook quickly becomes a chore as you scroll past long blocks of unintelligible base-64 gibberish.
This is a problem when working with long-lived, multiple-revision or (especially) multiple-coauthor projects. What can we do about this? First, it's worth mentioning the initial "I'll figure this out later" solution which has served many users sufficiently well for a while - if you're typically only working from one machine, and you just want to keep your notebooks vaguely manageable, you can get by for a long time by manually hitting Cell -> All Output -> Clear (followed by a Save) before you commit your notebooks. This wipes the slate clean with regards to cell outputs (plots, prints, whatver), so you'll need to re-run any computation next time you run the notebook.
The problems with this approach are that
A. it's manual, so you'll have to painstakingly open up every notebook you recently re-ran and clear it before you commit, and
B. it doesn't even fully solve the 'noise-in-the-diffs' problem, since every notebook also contains a 'metadata' section, which looks a bit like this:
{ "metadata": { "kernelspec": { "display_name": "Python 2", "language": "python", "name": "python2" }, "language_info": { "codemirror_mode": { "name": "ipython", "version": 2 }, "file_extension": ".py", "mimetype": "text/x-python", "name": "python", "nbconvert_exporter": "python", "pygments_lexer": "ipython2", "version": "2.7.12" } }
Note the metadata section is effectively a blank slate, and has a myriad of possible uses, but for most users it will just contain the above. This is useful for checking a previously run notebook, but is mostly unwanted information when checking-in files to a multi-user project where everyone's using a slightly different Python version - it just generates more diff-noise.
Possible Pythonic solutions
nbdime - an nbformat diff-GUI
We clearly need some tooling, and there are some Python projects out there trying to address exactly this problem. First, it's worth mentioning nbdime, which picks up the ball from where the (now defunct) nbdiff project left off and attempts to provide “content-aware” diffing and merging of Jupyter notebooks - a meld (GUI) diff-tool equivalent for the nbformat, if you will. I think nbdime has the potential to be a really good beginner-friendly, general purpose notebook-handling tool and I want to see it succeed. However; it's currently somewhat of a beta, and more importantly it only fills one role in the notebook editing toolbox - viewing crufty diffs. What I really want to do is automatically clear out all the cruft and minimize the diffs in the first place.
nbstripout - does what it says on the tin
A little searching then leads to nbstripout, which is a one-module Python script wrapping the nbformat processing functions, and adding some automagic for setting up your git config (on which more in a moment). This effectively automates the 'clear all output' manual process described above. However, this doesn't suit me for a couple of reasons; it leaves in that problematic 'metadata' section and also it's **slowww**. Running a script manually and expecting a short delay is fine, but we're going to integrate this into our git setup. That means it will run every time we hit git diff! One of the few things I love about git is that it's typically blazing fast; so a delay of nearly a fifth of a second every time I try to interact with it gets old pretty quickly:
time nbstripout 01-parsing.ipynb real 0m0.174s user 0m0.152s sys 0m0.016s
(Note, this is a small notebook-file, on a fairly beefy laptop with an SSD). This not a criticism of nbstripout so much as an inherent flaw in using Python for low-latency tasks - that cold-startup overhead on the CPython interpreter is a killer. (Which in turn harks back to ancient history of mercurial vs git!)
Enter jq
Fortunately, we have another option (thanks to Jan Schulz for the tip-off on this). Since the nbformat is just JSON, we can make use of jq, 'a lightweight and flexible command-line JSON processor' ('sed for JSON data'). There's a modicum of set-up overhead as jq has its very own query / filter language, but the documentation is good and the hard work has been done for you already. Here's the jq invocation I'm currently using:
jq --indent 1 \ ' (.cells[] | select(has("outputs")) | .outputs) = [] | (.cells[] | select(has("execution_count")) | .execution_count) = null | .metadata = {"language_info": {"name":"python", "pygments_lexer": "ipython3"}} | .cells[].metadata = {} ' 01-parsing.ipynb
Each line inside the single-quotes defines a filter - the first selects any entries from the 'cells' list, and blanks any outputs. The second resets any execution counts. The third wipes the notebook metadata, replacing it with the minimum of required information for the notebook to still run without complaints [*] and work correctly when formatted with nbsphinx. The fourth filter-line,
.cells[].metadata = {}
is a matter of preference and situation - in recent versions of Jupyter every cell can be marked hidden / collapsed / write-protected, etc. I'm not interested in that metadata usually but of course you may want to keep it for some projects.
We now have a fully stripped-down notebook that should contain only the common information needed to execute with whatever local Python installation is available (assuming Python2/3 compatibility, correctly set-up library installs and all the rest).
Note you'll need jq version 1.5 or greater, since the --indent option was only recently implemented and is necessary to conform with the nbformat. Fortunately that should only be a small binary-download away, even if you're on ancient linux or OSX.
That's a bit of a handful to type, but you can set it up as an alias in your .bashrc with a bit of careful quotation-escaping:
alias nbstrip_jq="jq --indent 1 \ '(.cells[] | select(has(\"outputs\")) | .outputs) = [] \ | (.cells[] | select(has(\"execution_count\")) | .execution_count) = null \ | .metadata = {\"language_info\": {\"name\": \"python\", \"pygments_lexer\": \"ipython3\"}} \ | .cells[].metadata = {} \ '"
Which can then be used conveniently like so:
nbstrip_jq 01-parsing.ipynb > stripped.ipynb Not only does this give us full control to wipe that pesky metadata, it's pretty damn quick, taking something like a tenth of the time of nbstripout in my (admittedly ad-hoc) testing:
nbstrip_jq 01-parsing.ipynb # (JSON contents omitted) real 0m0.015s user 0m0.008s sys 0m0.004s
Automation: Integrating with git
So we're all tooled up, but the question remains - how do we get git to run this automatically for us? For this, we dive into 'gitattributes' functionality, specifically the filter section. This describes how to define 'clean' and 'smudge' (reverse of clean) filters, which are operations that transform our data as it is checked in or out of the git-repository, so that (for example) our notebook-output cells are always stripped away from the JSON-data before it's added to the git repository:
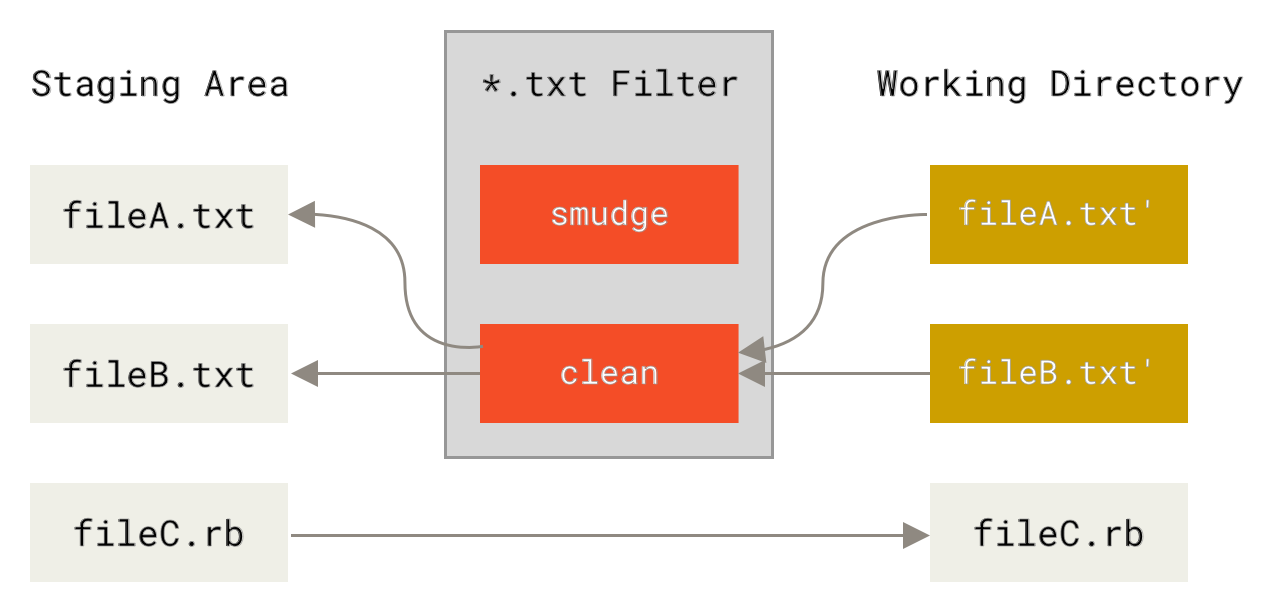
In the general case you can also define a smudge-filter to take your repository contents and do something with it to make it local to your system, but we'll not be needing that here - we'll just use the cat command as a placeholder. The easiest way to explain how to configure this is with an example. Personally, I want notebook-cleaning behaviour to be the default across all my git-repositories, so I have the following entries in my global ~/.gitconfig file:
[core] attributesfile = ~/.gitattributes_global [filter "nbstrip_full"] clean = "jq --indent 1 \ '(.cells[] | select(has(\"outputs\")) | .outputs) = [] \ | (.cells[] | select(has(\"execution_count\")) | .execution_count) = null \ | .metadata = {\"language_info\": {\"name\": \"python\", \"pygments_lexer\": \"ipython3\"}} \ | .cells[].metadata = {} \ '" smudge = cat required = true
And then in ~/.gitattributes_global:
*.ipynb filter=nbstrip_full
(Note, once you've defined your filter you can just as easily assign it to files in a repository specific .gitattributes file if you prefer a fine-grained approach.)
That's it! You're all set to go version control notebooks like a champ! Well, almost.
Getting started and gotchas
Note that we're into git-powertool territory here, so things might be a little less polished compared to the (cough) usual intuitive git interface you're used to.
To start off with, assuming a pre-existing set of notebooks, you'll want to add a 'do-nothing' commit, where you simply pull in the newly-filtered versions of your notebooks and trim out any unwanted metadata. Just git add your notebooks, noting that you may need to touch them first, so git picks up on the timestamp-modification and actually looks at the files for changes. Then,
to see the patch removing all the cruft. Commit that, then go ahead, run your notebooks, leave uncleaned outputs all over the place. Unless you change the actual code-cell contents, your git diff should be blank!
Great. Except. If you have executed a notebook since your last commit, git status may show that file as 'modified', despite the fact that when you git diff, the filters go into action and no differences-to-HEAD are found. So you have to 'tune out' these false-positive modified flags when reading the git-status. Another issue is that if you use a diff-GUI such as meld, then beware: unlike git diff, git difftool will not apply filters to the working directory before comparing with the repo HEAD - so your command-line and GUI diffs have suddenly diverged! The logic behind this difference in behaviour is that GUI programs give the option to edit the local working-copy directly, as discussed at length in this thread. This has clearly caught out others before.
If they bother you, these false-positives and diff-divergences can easily be resolved by manually applying the jq-filters before you run your diffs. For convenience, my ~/.bashrc also defines the following command to apply the filters to all notebooks in the current working directory:
function nbstrip_all_cwd { for nbfile in *.ipynb; do echo "$( nbstrip_jq $nbfile )" > $nbfile done unset nbfile }
Addtionally, let me note that clean/smudge filters often do not play well with rebase operations. Things get very confusing if you try to rebase across commits before / after applying a clean-filter. The simplest way to work around this is to simply comment out the relevant filter-assignment line in .gitattributes_global while performing a rebase, then uncomment it when done.
As a parting note, if you also choose to configure your gitattributes globally, you may want to know how to 'whitelist' notebooks in a particular repository (for example, if you're checking-in executed notebooks to a github-pages documentation branch). This is dead easy, just add a local .gitattributes file to the repository and 'unset' the filter attribute, like so:
Or you could replace the *.ipynb with a path to a specific notebook, etc.
Hope that helps! Comments or corrections very welcome via Twitter.
from Hacker News https://ift.tt/2u0NtnH
No comments:
Post a Comment
Note: Only a member of this blog may post a comment.